4 Usage
Sample code demonstrating the use of pyPAHdb is shown below, which has been
included in the pyPAHdb package as examples/example_tbl.py. The example
makes use of the bundled sample_data_NGC7023.tbl spectrum.
import importlib_resources
from pypahdb.decomposer import Decomposer
from pypahdb.observation import Observation
if __name__ == "__main__":
# Spitzer sample data (IPAC table).
file_path = importlib_resources.files("pypahdb")
data_file = file_path / "resources/sample_data_NGC7023.tbl"
# Construct an Observation object.
obs = Observation(data_file)
# Pass the Observation's spectrum to Decomposer, which performs the fit.
pahdb_fit = Decomposer(obs.spectrum)
# Save the fit to disk, both as a PDF and FITS file.
pahdb_fit.save_pdf("NGC7023_pypahdb_tbl_example.pdf", domaps=False)
pahdb_fit.save_fits("NGC7023_pypahdb_tbl_example.fits", header=obs.header)
4.1 Details
Let’s briefly explore what the code above is doing.
First the Decomposer and Observation classes are imported from
pyPAHdb. The``importlib_resources`` package is used to locate and load the
provided sample data.
import importlib_resources
from pypahdb.decomposer import Decomposer
from pypahdb.observation import Observation
if __name__ == "__main__":
# Spitzer sample data (IPAC table).
file_path = importlib_resources.files("pypahdb")
data_file = file_path / "resources/sample_data_NGC7023.tbl"
The included example IPAC table, sample_data_NGC7023.tbl, contains the
astronomical PAH spectrum shown below:
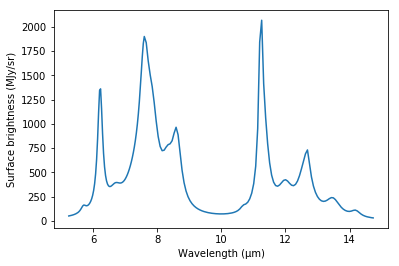
The file is read with the Observation class by instantiating it and passing
the path as its argument:
# Construct an Observation object.
obs = Observation(data_file)
Note that that the Observation class is able to handle a variety of
file-formats, not just IPAC tables.
The decomposition is performed by creating a Decomposer-instance and
passing it obs.spectrum as its only positional argument.
.. code-block:: python
# Pass the Observation’s spectrum to Decomposer, which performs the fit. pahdb_fit = Decomposer(obs.spectrum)
The Decomposer class has two methods for producing output. On for saving
to PDF; save_pdf, the other for saving to FITS, save_fits. Both
methods require the filename to write to as their first positional argument.
# Save the fit to disk, both as a PDF and FITS file.
pahdb_fit.save_pdf('NGC7023_pypahdb_tbl_example.pdf', domaps=False)
pahdb_fit.save_fits('NGC7023_pypahdb_tbl_example.fits', header=obs.header)
Note that header=obs.header is explicitly passed to save_fits, but
can be set arbitrary, i.e., it is possible to provide a customized the header.